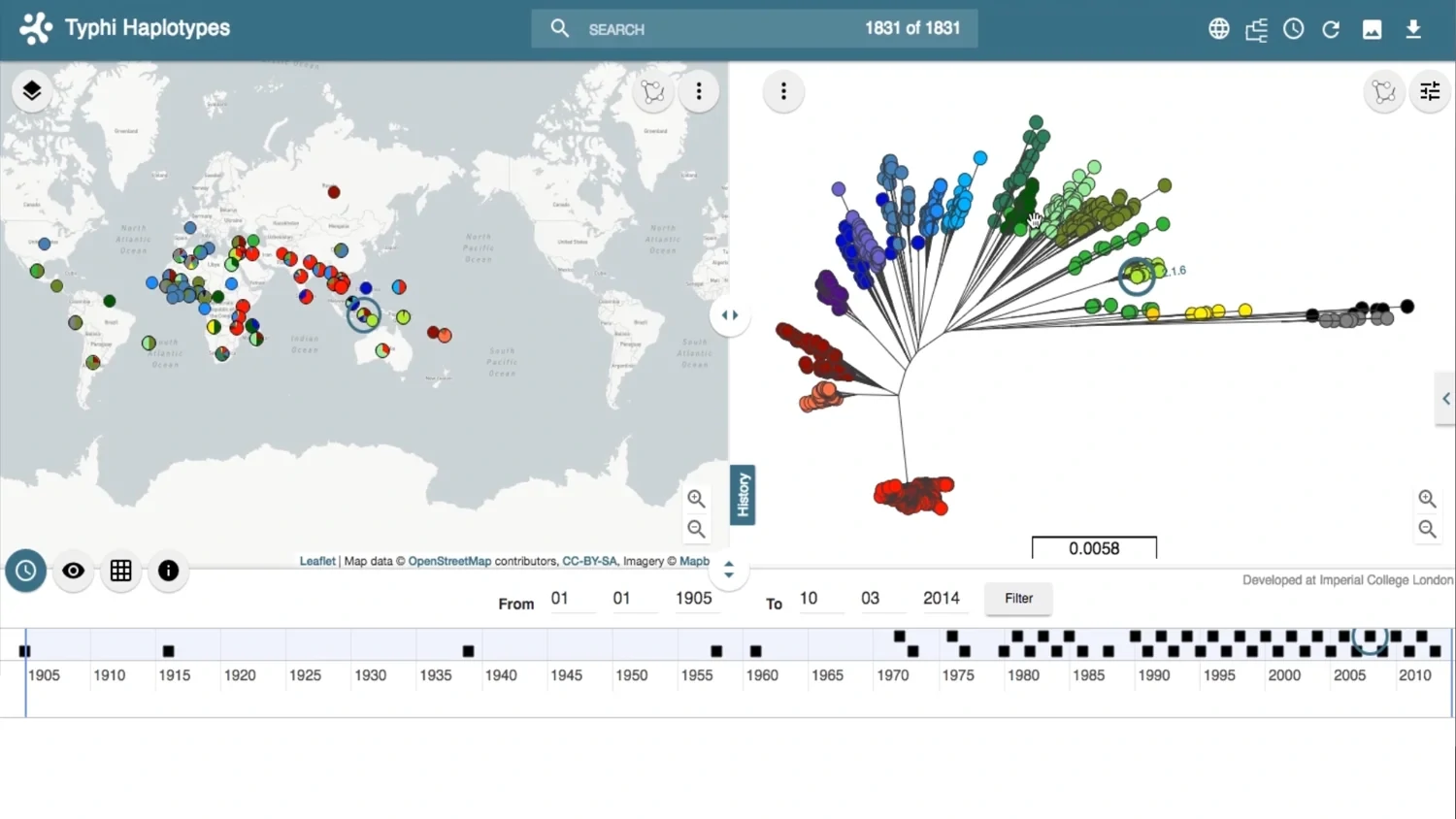
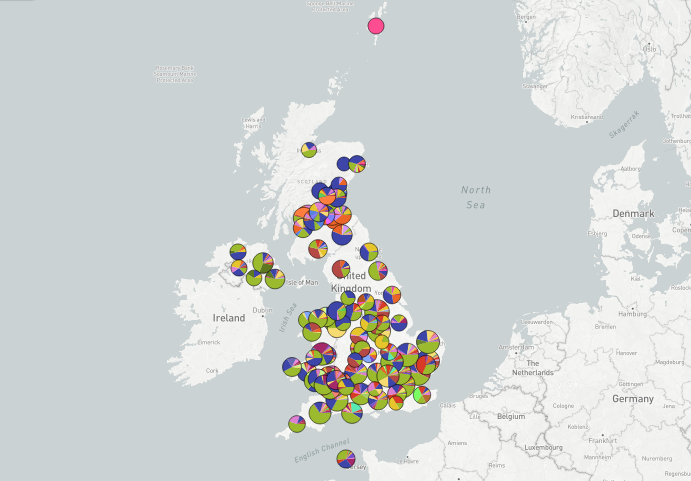
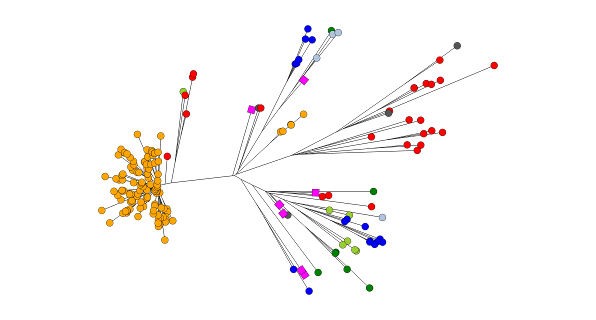
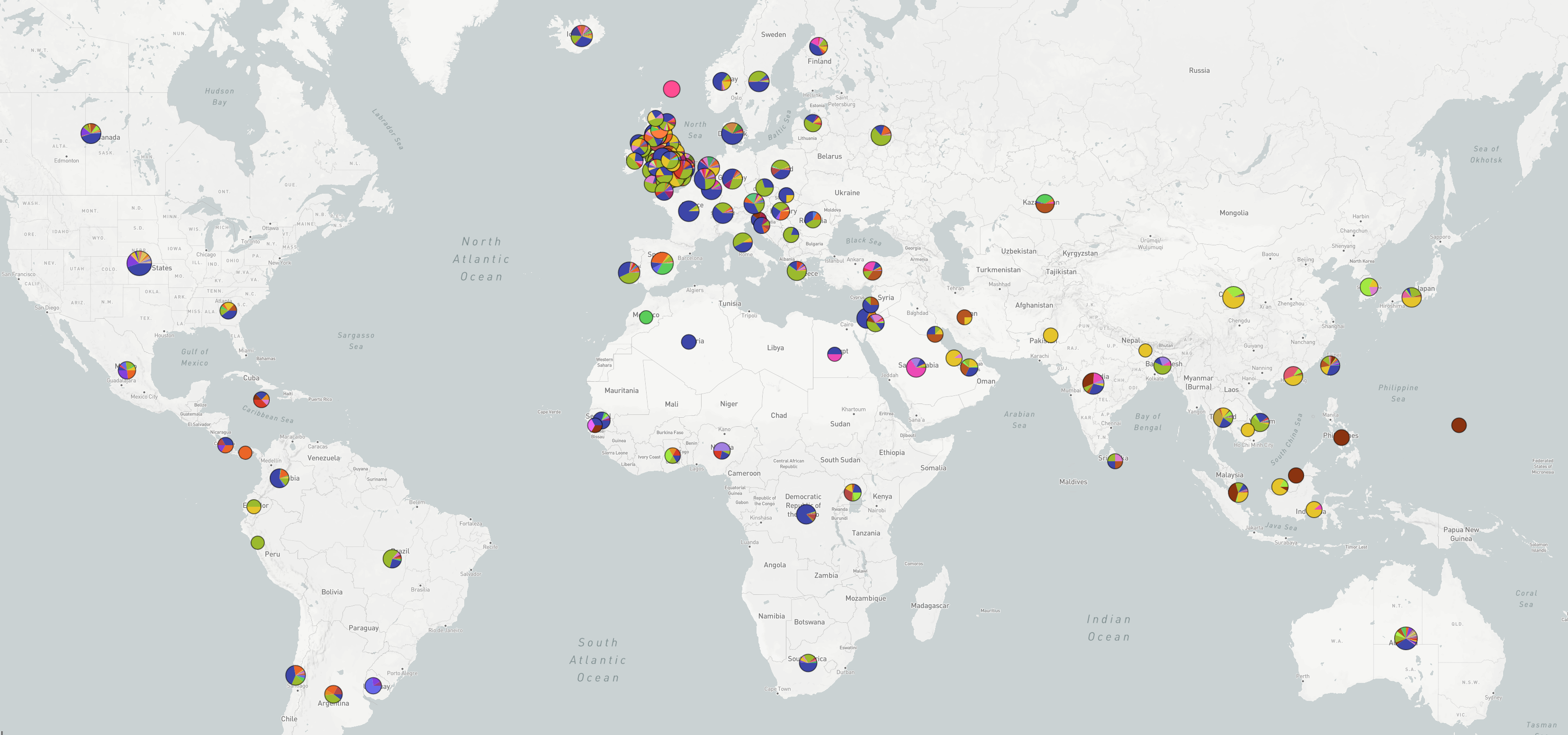
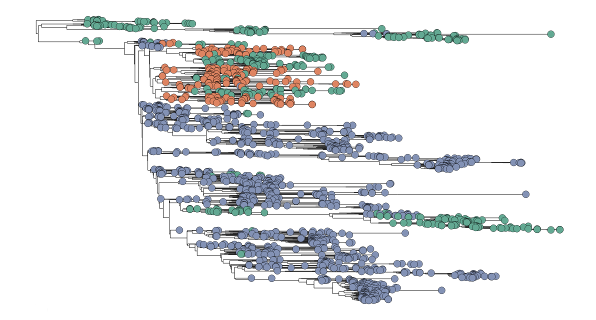
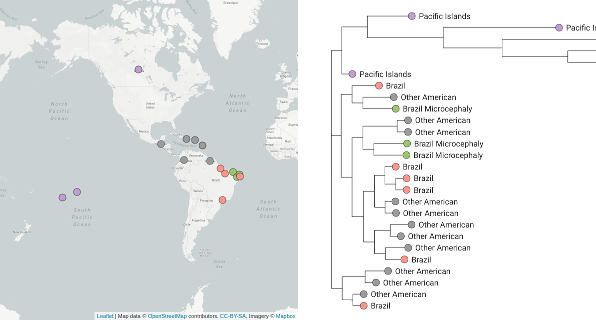
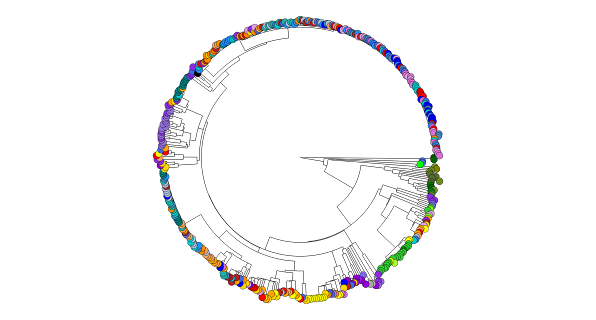
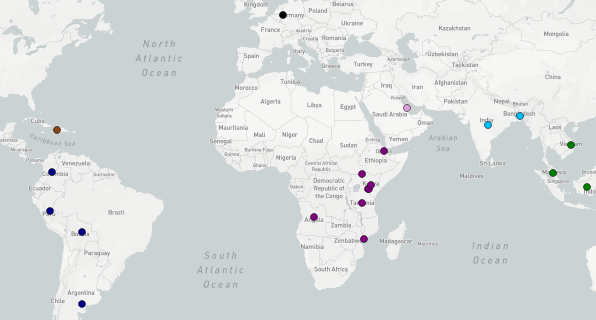
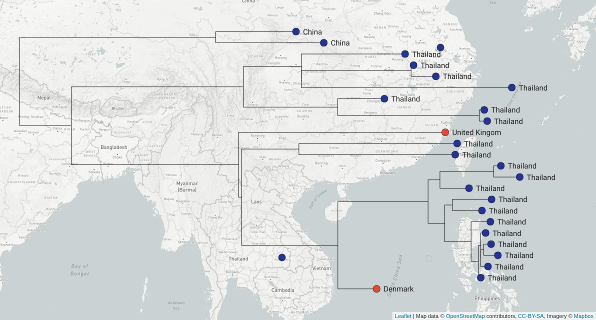
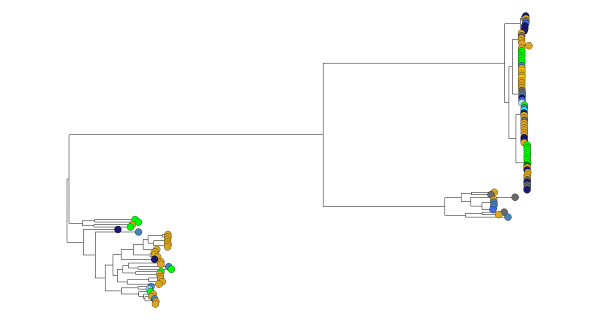
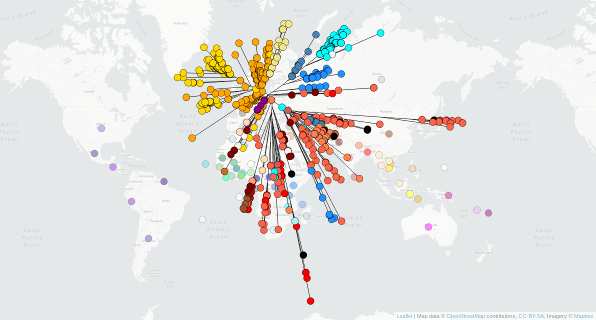
Tracing the spread of carbapenem-resistant Klebsiella pneumoniae
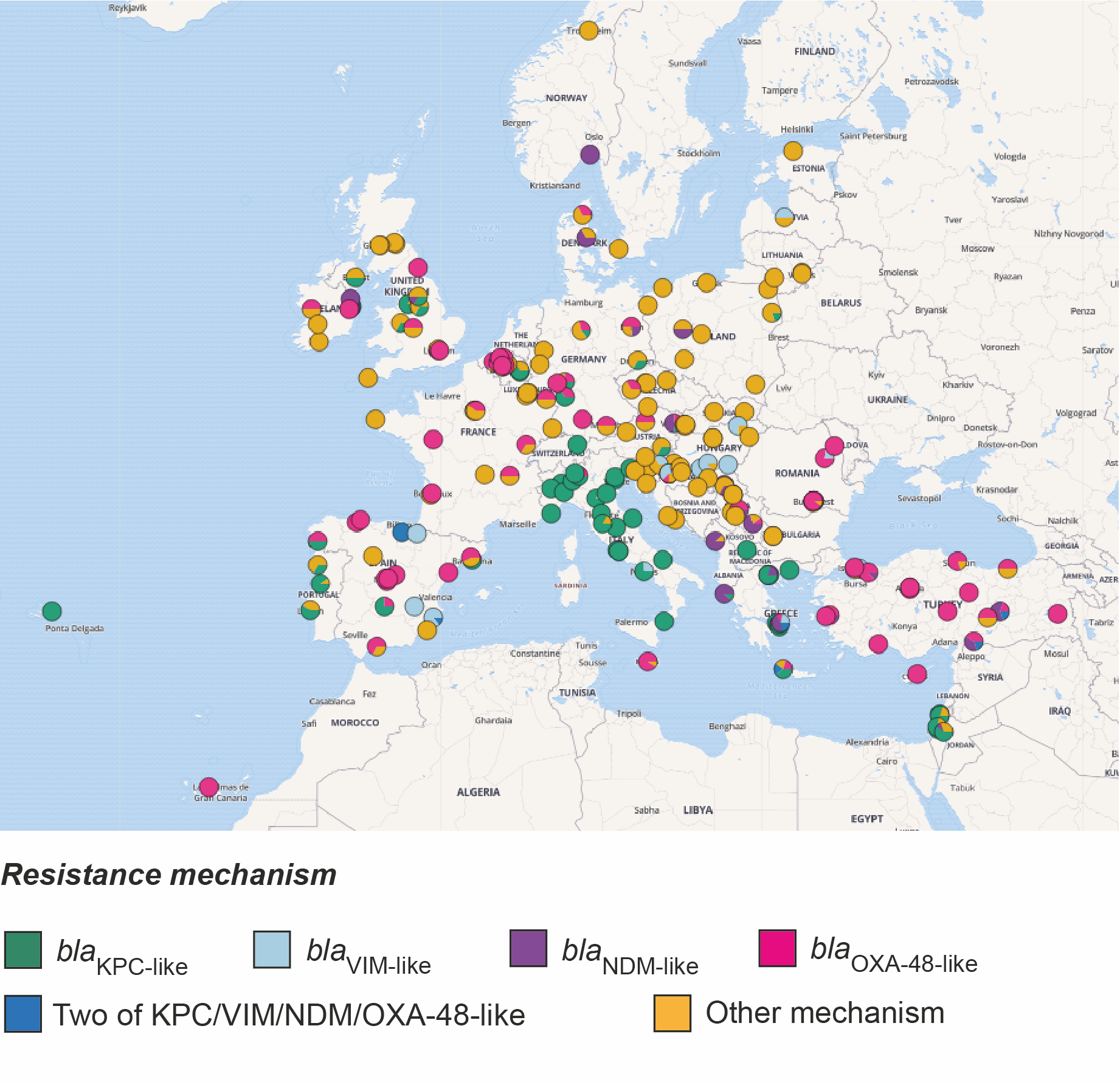
Epidemic of carbapenem-resistant Klebsiella pneumoniae in Europe is driven by nosocomial spread. Nature Microbiology 2058-5276. (doi:10.1038/s41564-019-0492-8)
https://www.nature.com/articles/s41564-019-0492-8